Replication and nuclear Dynamics
Research Project
Our research focus is to decipher how cell identity and gene expression are regulated during cell division and understand the implication of these molecular processes in various human diseases. We focus on a particular moment of the cell cycle: the S phase, that is when the genetic material is duplicated. During DNA replication, chromatin undergoes significant structural alterations. The progression of the replication machinery forces the unwinding of the two DNA strands, creating topological constrains and promoting the eviction of nucleosomes and chromatin binding proteins ahead of the replication fork. This leads to changes in chromatin accessibility and a dilution of the epigenetic information that can alter gene expression regulation.
Given that chromatin restoration can span hours, in our group we want to analyze the impact of all these chromatin rearrangements in key biological processes focusing on the following lines of research:
1. Explore how chromatin replication alters RNA polymerase II (RNAPII) activity
2. Investigate how chromatin replication modifies RNA synthesis
3. Unveil how these molecular mechanisms impact on human diseases linked to cell division such as cancer
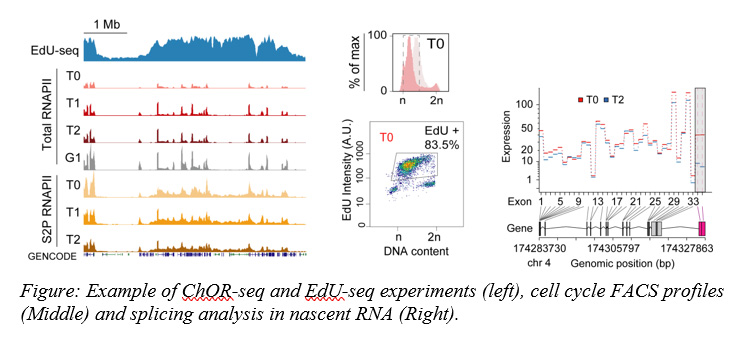
To achieve our research goals, we use human and mouse cell cultures and employ cutting-edge molecular biology techniques coupled with high-throughput sequencing (such as ChOR-seq, ChIP-seq, RNA-seq, ChrRNA-seq, EdU-seq) to explore newly replicated chromatin and newly transcribed RNAs.
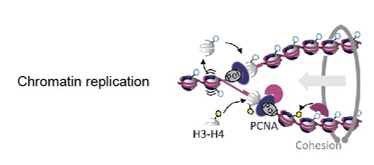
Our findings have revealed that after chromatin replication both chromatin rearrangements and transcription-replication conflicts produced during normal DNA replication alter RNAPII activity and lead to transient quantitative and qualitative changes in RNA synthesis. Understanding how all these events are regulated will be critical to understand physiological and pathological human processes linked to cell division and will help us to design new and more effective treatments to human diseases such as cancer.
If you are interested in joining our group as a master student, PhD student or Postdoc, please send us your CV to: cristina.gonzalez@cabimer.es
Funding:

- Bruno F, Coronel-Guisado C and Gonzalez-Aguilera C (2024) Collisions of RNA polymerases behind the replication fork promote alternative splicing in newly replicated chromatin. Mol Cell. 84(2):221-233.e6 doi:10.1016/j.molcel.2023.11.036. (Corresponding Author). Highlighted in a Preview in Mol Cell: Werner M and Hamperl S (2024) Mol Cell. 84(2):186-188. doi: 10.1016/j.molcel.2023.12.029.
- Petryk N *, Reveron-Gomez N*, Gonzalez-Aguilera C*, Dalby M, Andersson R, Groth A. (2021) Genome-wide and sister chromatid-resolved profiling of protein occupancy in replicated chromatin with ChOR-seq and SCAR-seq. Nat Protocol. 16(9) 4446-4493 *EQUAL CONTRIBUTION. IF: 10,42.
- Martin-Broto J, CruzJ, PenelN, LecesneA, HindiA, LunaP, MouraDS, BernabeuD, de AlavaE, Lopez-GuerreroJA, DopazoJ, Peña-ChiletM, GutierrezA, ColliniP, KaranianM, RedondoA, Lopez-PousaA, GrignaniG, Diaz-MartinJ, MarcillaD, Fernandez-SerraA, Gonzalez-Aguilera C, Casali PG, Blay JY and Stacchiotti (2020) Pazopanib for treatment of typical solitary fibrous tumour: A multicentre single-arm, Phase II trial. The Lancet Oncology (In press).
- Reverón-Gómez N*, González-Aguilera C*, Stewart-Morgan KR*, Petryk N, Flury V, Graziano S, Johansen JV, Jacobsen JS, Alabert C and Groth A. (2018) Accurate recycling of parental histones reproduces the histone modification landscape during DNA replication. Cell. 72(2):239-249.e5 doi: 10.1016/j.molcel.2018.08.010. *EQUAL CONTRIBUTION. Zaugg J: F1000Prime recommendation of Reverón-Gómez N et al., Mol Cell. 2018 72(2):239-249.e5 in F1000Prime, 31 Oct 2018 doi: 10.3410/f.733853367.793552123.
- Huang H, Strømme CB, Saredi G, Hödl M, Strandsby A, González-Aguilera C, Chen S, Groth A, Patel DJ (2015) A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 22(8):618-26.
- Tena JJ*, González-Aguilera C*, Fernández-Miñán A, Wittbrodt J, Gómez-Skarmeta JL, Martínez-Morales JR. (2014) Comparative epigenomics in distantly related teleost species identifies conserved cis-regulatory nodes active during the vertebrate phylotypic period. Genome Res. DOI: 1101/gr.163915.113 *Equal contribution.
- González-Aguilera C, Ikegami K., Ayuso C., de Luis A., Íñiguez M., Cabello J., Lieb JD. and Askjaer P. (2014) Genome-wide analysis links emerin to neuromuscular junction activity in C. elegans. Genome Biol. Feb 3;15(2):R21.
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D., Kalck V., Meister P, Askjaer P, and Gasser SM. (2012) Step-wise methylation of histone H3K9 positions chromosome arms at the nuclear envelope in C. elegans embryos. 150, 934-47.
- González-Aguilera, C., Tous, C., Babiano R., de la Cruz J., Luna, R:, and Aguilera, A (2011) Nab2 functions in the metabolism of RNA driven by polymerases II and III. Biol. Cell. 22(15):2729-40.
- Gonzalez-Aguilera, C., Tous, C., Gomez-Gonzalez, B., Huertas, P., Luna, R., and Aguilera, A. (2008) The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Biol. Cell 19(10), 4310-4318.
Complete list of publications:
SCOPUS ID https://www.scopus.com/authid/detail.uri?authorId=24070509800
ORCID ID https://orcid.org/0000-0003-4845-5744