e-mail: jonathan.martinez@cabimer.es
ORCID ID: 0000-0001-5809-065X
Research Line:
- Understanding the physiopathological role of the non-canonical, extralysosomal functions of lysosomal proteases and its connection with inflammation.
e-mail: jonathan.martinez@cabimer.es
ORCID ID: 0000-0001-5809-065X
Research Line:
Currently I am an independent researcher awarded an EMERGIA fellresearch grant (Junta de Andalucia) started on September 2023, that allows me to continue with my own line of research on unravelling the physiopathological role of the non-canonical, extralysosomal functions of lysosomal proteases and its connection with pro-inflammatory conditions. Previously, I was also awarded a Ramon y Cajal Fellow (call 2021).
My scientific career has been focused in the study of the lysosomal compartment and its connection with physiopathology, both through their well-characterised canonical, intralysosomal activities, but also through their non-canonical, extralysosomal functions. During my scientific career I have worked in several scientific projects both at national and international level (European Research Council, Welcome Trust, Medical Research Council, Spanish Ministry of Science), published a number of scientific articles in high-impact factor journals (i.e., Science Immunology, Immunity, Blood, Nature Communications, Nature Structural and Molecular Biology, Cell Reports, eLIFE, among others). I have presented my work at more than 20 national and international conferences, been invited to several talks at international conferences and awarded several fellowships.
During my Postdoctoral work at the University of Dundee (UoD, UK), I led a project about the characterisation of a novel STAT3-mediated pathway of lysosomal homeostasis in which I designed and executed all the experiments, leading to a first and corresponding author paper in Nature Communications in 2018. This allowed me to secure a position as Senior Research Associate (SRA) at the UoD in which I developed my own, independent line of research.
Importantly, my stay in the United Kingdom has allowed me to stablish collaborations with some of the world leading experts in the fields of Immunology (Prof. Colin Watts (UK); Prof. Doreen Cantrell (UK), Dr Suman Mitra (FR)), Lysosomal Biology (Prof. Thomas Reinheckel (DE); Prof. Valeria Poli (IT)), Bioinformatics (Prof. Angus Lamond (UK); Dr Majid Kazemian (USA)) and Cytokine Biology (Dr Ignacio Moraga (UK)). This excellent network of collaborators provides excellent support in the development of my scientific career, allowing me to explore lysosomal biology from different scientific perspectives.
Team members:
Jonathan Martinez Fabregas (Project leader)
Ryan Conesa Bakkali (PhD Student)
Previous positions:
Lysosomes, initially described by De Duve in 1955 were for a long time considered only the recycling plants of cells but are now recognised as participating in multiple aspects of cell physiology. Lysosomes play a central role in the regulation of the immune system—(I) Toll-like receptor signalling, (II) generation of immunological informationand (III) cytotoxic killing—but also in the normal physiology of the cells—(IV) mTOR pathway, (V) autophagy and (VI) lysosomal homeostasis (LH). Furthermore, lysosomal hydrolases have started to be linked to the regulation of physiological processes through their extralysosomal functions—(VII) regulation of cell division, (VIII) LM-PCD, (IX) neurotoxicity and (X) cell differentiation (Figure 1).
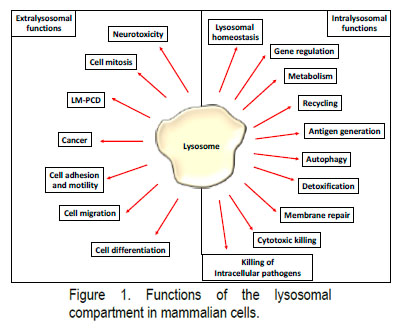
The broad range of physiological processes regulated by lysosomes helps us to rationalise why lysosomal dysfunction is involved at the onset of several human pathologies such as Lysosomal Storage Diseases (LSDs), autoinflammatory, autoimmune and neurodegenerative diseases, and cancer.
However, the ability of these lysosomal hydrolases to control physiological processes occurring in the cytosolic and the nuclear compartment has started to emerge. Indeed, lysosomal proteases are nowadays accepted to play extralysosomal functions during the onset and progression of Lysosomal-Mediated Programmed Cell Death (LM-PCD), during cell division, in neurotoxicity and during cell differentiation, etc. Thus, a deeper understanding of the signalling pathways involved in the regulation of lysosomal hydrolytic capacity and the extralysosomal roles played by these lysosomal hydrolases could help us to gain a deeper understanding of lysosomal biology and furthermore provide new tools to ameliorate the symptoms associated with these diseases.
On one hand, the mechanism by which lysosomal gene expression is regulated was in part elucidated by the discovery of the role played by the mTOR-TFEB axis which links lysosomal biogenesis and autophagy. However, as recently shown by our work and others the cytokine-STAT3 signalling hub also contributes to the regulation of the lysosomal compartment. However, the molecular mechanisms of the cytokine-mediated regulation of lysosomes remain unsolved. On this regard, our work is focused in characterising the pleiotropic role played by pro- and anti-inflammatory cytokines regarding the regulation of the lysosomal compartment in immune cells (Figure 2).
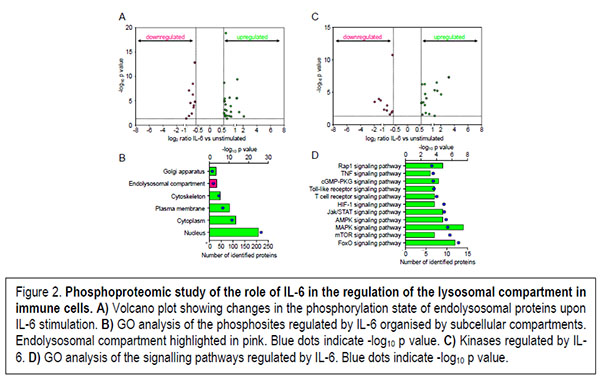
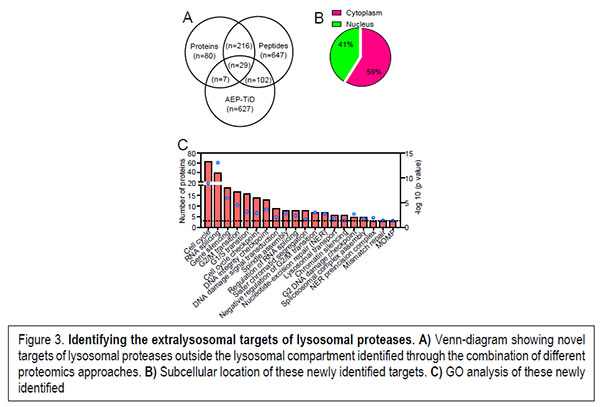
On the other hand, the extralysosomal role played by lysosomal hydrolases has recently started to be elucidated by the identification of physiological processes where these hydrolases play essential roles. However, a high-throughput approach aimed at the identification of the extralysosomal targets of these hydrolases that could help to us to picture the role played by these hydrolases upon release from the lysosomal compartment is still missing. Our preliminary work is allowing us to start unveiling the biological processes regulated by these lysosomal proteases when located in the nuclear compartment under pathological conditions (Figure 3).
Projects as PI:
Articles:
A complete list of publications can be obtained at: