Genome Instability & Cancer
 Our research interests are to identify and understand the causes and mechanisms of genome instability, a hallmark of cancer cells. To preserve genome integrity, DNA replication is necessarily coordinated with transcription, sister chromatid cohesion and chromatin assembly and remodeling. In addition, cells need to deal with DNA damage, not only originated by exogenous and endogenous agents but also by transcription and non-B DNA structures such as R loops. Failure to do so may lead to DNA breaks, rearrangements and mutations that can cause loss of viability and defects in cell proliferation. As a consequence, errors in replication and the DNA Damage Response (DDR) can result in aging, cancer predispositions and genetic abnormalities. Our research, performed in human cells and the model organism Saccharomyces cerevisiae, pursues the following goals:
Our research interests are to identify and understand the causes and mechanisms of genome instability, a hallmark of cancer cells. To preserve genome integrity, DNA replication is necessarily coordinated with transcription, sister chromatid cohesion and chromatin assembly and remodeling. In addition, cells need to deal with DNA damage, not only originated by exogenous and endogenous agents but also by transcription and non-B DNA structures such as R loops. Failure to do so may lead to DNA breaks, rearrangements and mutations that can cause loss of viability and defects in cell proliferation. As a consequence, errors in replication and the DNA Damage Response (DDR) can result in aging, cancer predispositions and genetic abnormalities. Our research, performed in human cells and the model organism Saccharomyces cerevisiae, pursues the following goals:
- to identify the mechanisms by which cells counteract R-loops and transcription-replication conflicts
- to resolve the mechanism by which mRNA biogenesis factors control genome integrity
- to decipher the role of chromatin and RNA in replication fork integrity and DSB repair
- to use new determinants of transcription-dependent replication stress in cancer diagnostics and therapy.
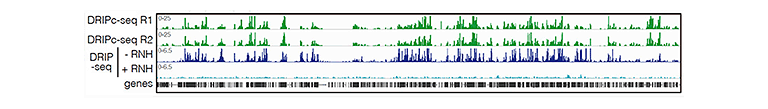
Research lines

- R-loops and transcription-replication conflicts in genome instability
We investigate the mechanisms by which cells solve transcription-replication conflicts and the role of specific DDR and chromatin factors such as BRCA2, FANCD2, SWI/SNF, FACT, SIN3 and others. We focus on the impact of RNA-DNA hybrids in these conflicts and how they lead to chromosome breakage. - Role of mRNP-biogenesis factors in genome integrity
We study the mechanisms of action of yeast and human RNA-metabolic factors, such as THO, THSC/TREX-2, UAP56/DDX39B, ALY-REF and others during transcription and RNA export in relation to the nuclear structure to understand their involvement in DNA-RNA hybrid homeostasis and how they control genome integrity. - Role of chromatin and RNA at broken replication forks
We study the role of chromatin remodelers, recombinational DSB factors, DNA-RNA hybrid and RNA-metabolic factors in the restart of broken forks and in the repair of replication-born double-strand breaks (DSBs) via sister-chromatid recombination.
- RNA-mediated replication stress in cancer
We investigate a possible connection between R-loops and cancer cells, via the role of tumor suppressors and anti-tumor drugs in R-loop-mediated genome instability and their impact on cell proliferation and genome integrity.
For PhD STUDENT and POSTDOC APPLICATIONS send motivation letter, CV and e-mail of 3 referees to genetic.instability@cabimer.es
Curriculum Vitae Andrés Aguilera
Twitter:
Funding:






Full list of publications:
Recent selected publications:
San Martin-Alonso M, Soler-Oliva ME, García-Rubio M, García-Muse T, Aguilera A. 2021. Harmful R-loops are prevented via different cell cycle-specific mechanisms. Nat. Commun. 22;12(1):4451.
Ortega P, Mérida-Cerro JA, Rondón AG, Gómez-González B, Aguilera A. 2021. DNA-RNA hybrids at DSBs interfere with repair by homologous recombination. Elife 10:e69881.
Bayona-Feliu A, Barroso S, Muñoz S, Aguilera A. 2021. The SWI/SNF chromatin remodeling complex helps resolve R-loop-mediated transcription-replication conflicts. Nat. Genet. 53(7):1050-1063.
Sessa G, Gómez-González B, Silva S, Pérez-Calero C, Beaurepere R, Barroso S, Martineau S, Martin C, Ehlén Å, Martínez JS, Lombard B, Loew D, Vagner S, Aguilera A*, Carreira A*. 2021. BRCA2 promotes DNA-RNA hybrid resolution by DDX5 helicase at DNA breaks to facilitate their repair. EMBO J. 40(7):e106018.
Pérez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Muñoz S, González-Basallote VM, Sung P, Aguilera A. 2020. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 34(13-14):898-912. Erratum in: Genes Dev. 2021 35(7-8):573.
Barroso S, Herrera-Moyano E, Muñoz S, García-Rubio M, Gómez-González B, Aguilera A. 2019. The DNA damage response acts as a safeguard against harmful DNA-RNA hybrids of different origins. EMBO Rep. 20(9):e47250.
García-Muse T, Aguilera A. 2019. R Loops: From Physiological to Pathological Roles. Cell 179(3):604-618. [Review]
Gómez-González B, Aguilera A. 2019. Transcription-mediated replication hindrance: a major driver of genome instability. Genes Dev. 33(15-16):1008-1026. [Review]

 Assist. Prof. Belén Gómez González
Assist. Prof. Belén Gómez González Assit Prof. Emilia Herrera-Moyano
Assit Prof. Emilia Herrera-Moyano Assoc. Prof. Rosa Luna
Assoc. Prof. Rosa Luna






