Cell division control
The research in our group aims to shed light on the molecular mechanisms that control cell division and ensure the proper distribution of the genome during this process. Problems with the partition of the genetic material during cell division can give rise to conditions like aneuploidy, an alteration of the normal number of chromosomes in the cell that is a hallmark of cancer and a number of different genetic diseases. Advances in our knowledge about the mechanisms that regulate this process are thus of pivotal importance to better understand the development of such diseases. With this general objective, the main research lines in our laboratory are:
1- Regulation of cell cycle progression and analysis of the surveillance mechanisms that ensure the fidelity in the distribution of the genome during this process:
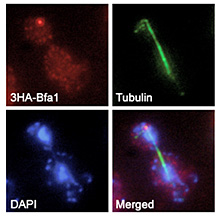 The cells have developed a number of surveillance mechanisms to verify the integrity of the genetic material and its proper distribution during cell division, such as the DNA damage checkpoint (DDC), which detects lesions in the DNA and promotes their repair, or the spindle assembly checkpoint (SAC), which ensures that every chromosome has been attached to the mitotic spindle. In asymmetric cell divisions, additional surveillance mechanisms also control the orientation of the spindle. This was originally found in the yeast Saccharomyces cerevisiae. In this organism, due to its particular pattern of cell division by budding, the spindle must be perpendicularly aligned with respect to the division axis, thereby ensuring an equal partitioning of the chromosomes between the mother and the newly-generated daughter cell. The spindle position checkpoint (SPOC) avoids that budding yeast cells with a misaligned spindle exit from mitosis. We are extremely interested in understanding the molecular mechanisms that control the activity of the DDC, the SAC and the SPOC, therefore maintaining the correct ploidy during cell division.
The cells have developed a number of surveillance mechanisms to verify the integrity of the genetic material and its proper distribution during cell division, such as the DNA damage checkpoint (DDC), which detects lesions in the DNA and promotes their repair, or the spindle assembly checkpoint (SAC), which ensures that every chromosome has been attached to the mitotic spindle. In asymmetric cell divisions, additional surveillance mechanisms also control the orientation of the spindle. This was originally found in the yeast Saccharomyces cerevisiae. In this organism, due to its particular pattern of cell division by budding, the spindle must be perpendicularly aligned with respect to the division axis, thereby ensuring an equal partitioning of the chromosomes between the mother and the newly-generated daughter cell. The spindle position checkpoint (SPOC) avoids that budding yeast cells with a misaligned spindle exit from mitosis. We are extremely interested in understanding the molecular mechanisms that control the activity of the DDC, the SAC and the SPOC, therefore maintaining the correct ploidy during cell division.
Remarkably, in S. cerevisiae cells, and although the DDC, the SAC and the SPOC are triggered by different signals and in different stages of the cell cycle, all these checkpoints promote the inhibition of mitotic exit, which highlights the importance of the regulation of this cell cycle transition. Mitotic exit involves the disassembly of the mitotic spindle, the de-condensation of the chromosomes and the execution of cytokinesis, therefore leading to the final duplication of the cell. In S. cerevisiae, exit from mitosis is promoted by the Mitotic Exit Network (MEN), a signaling pathway that is initiated by the Tem1 GTPase. MEN signaling is blocked by the DDC, the SAC and the SPOC through the activation of Bfa1/Bub2, two proteins that together inhibit Tem1 activity. The regulation of mitotic exit, both during a normal cell cycle and after the activation of the mitotic checkpoints, is also a main research line developed in our laboratory.
2- Control of chromosome segregation by Aurora B kinase:
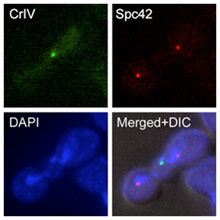 The elucidation of the mechanisms that facilitate the proper segregation of the chromosomes during cell division is extremely important to understand how cells maintain the correct ploidy during cell division. A main focus of our research group is the evaluation of the role of the Aurora B kinase in the control of this process. Aurora B is an essential protein that repairs erroneous chromosomal attachments that do not lead to the bi-orientation of sister chromatids. The accurate control of the activity of this kinase is crucial for the cells. The lack of Aurora B leads to severe chromosome segregation defects, aneuploidy and cell lethality. Interestingly, increased expression of this kinase also leads to aneuploidy, and it has been linked to different cancer types. In our laboratory, we are studying how the activity of Aurora B is controlled throughout the cell cycle, as well as how this kinase collaborates with the SAC in order to ensure the proper attachment of the chromosomes to the spindle. Finally, research in our group also tries to provide new insights into how the changes in the levels of Aurora B activity can determine problems during cell division.
The elucidation of the mechanisms that facilitate the proper segregation of the chromosomes during cell division is extremely important to understand how cells maintain the correct ploidy during cell division. A main focus of our research group is the evaluation of the role of the Aurora B kinase in the control of this process. Aurora B is an essential protein that repairs erroneous chromosomal attachments that do not lead to the bi-orientation of sister chromatids. The accurate control of the activity of this kinase is crucial for the cells. The lack of Aurora B leads to severe chromosome segregation defects, aneuploidy and cell lethality. Interestingly, increased expression of this kinase also leads to aneuploidy, and it has been linked to different cancer types. In our laboratory, we are studying how the activity of Aurora B is controlled throughout the cell cycle, as well as how this kinase collaborates with the SAC in order to ensure the proper attachment of the chromosomes to the spindle. Finally, research in our group also tries to provide new insights into how the changes in the levels of Aurora B activity can determine problems during cell division.
3- Generation of asymmetry during cell division:
Our group is also finally interested in the analysis of the mechanisms by which the cell generates asymmetry during cell division. S. cerevisiae is an ideal model organism where to study asymmetric cell divisions. Due to its particular way of division by budding, each cell division in this organism is intrinsically asymmetric, and the newly generated daughter cell differs from its mother in size, cellular composition, and replicative life span. Asymmetric cell divisions require the polarization of the cell along a predetermined axis, followed by the orientation of the mitotic spindle along this polarity axis and perpendicular to the division plane, this way ensuring the asymmetric distribution of polarization factors between the mother and the newly generated cell. Interestingly, the mitotic spindle is, in fact, an asymmetric structure in nature, especially at the level of the microtubule-organizing centers, from which the microtubules that constitute the spindle emanate. In our laboratory, we are trying to evaluate the role of spindle-associated asymmetries in the regulation of cell cycle progression, as well as the consequences that the problems during the establishment of these asymmetries determine on cell division, differentiation or aging. In higher eukaryotes, the stereotypical example of asymmetric cell divisions are stem cells. The asymmetric division of stem cells is a key mechanism to sustain the correct number of these cells in the different tissues. A reduction in the population of stem cells causes tissue disorganization, degeneration and aging. Conversely, an excessive stem cell number determines tissue hyperplasia and can contribute to the development of cancer. New insights into the mechanisms that regulate asymmetric cell divisions are extremely important to understand the origin of these problems.
a) Research articles:
– “Polo-like kinase acts as a molecular timer that safeguards the asymmetric fate of spindle microtubule-organizing centers”. Matellán L, Manzano-López J, Monje-Casas F. eLife (2020). 2(9): e61488. doi: 10.7554/eLife.61488.
– “Asymmetric inheritance of spindle microtubule-organizing centers preserves replicative lifespan”. Manzano-López J, Matellán L, Álvarez-Llamas A, Blanco-Mira JC, Monje-Casas F. Nature Cell Biology (2019). 21(8): 952-965. doi: 10.1038/s41556-019-0364-8.
– “Late rDNA condensation ensures timely Cdc14 release and coordination of mitotic exit signaling with nucleolar segregation”. de los Santos-Velázquez AI, de Oya IG, Manzano-López J, Monje-Casas F. Current Biology (2017). 27(21):3248-3263.e5. doi: 10.1016/j.cub.2017.09.028.
– “Dispensability of the SAC depends on the time window required by Aurora B to ensure chromosome biorientation”. Muñoz-Barrera M, Aguilar I, Monje-Casas F. PLoS One (2015). 10(12): e0144972. doi: 10.1371/journal.pone.0144972.
– “Increased Aurora B activity causes continuous disruption of kinetochore-microtubule attachments and spindle instability”. Muñoz-Barrera M, Monje-Casas F. Proc Natl Acad Sci USA (2014). 111(38): E3996-E4005. doi:10.1073/pnas.1408017111.
– “Inhibition of the Mitotic Exit Network in response to damaged telomeres”.
Valerio-Santiago M, de los Santos-Velázquez AI, Monje-Casas F. PLoS Genetics (2013). 9(10): e1003859. doi: 10.1371/journal.pgen.1003859.
– “Tem1 localization to the spindle pole bodies is essential for mitotic exit and impairs spindle checkpoint function”. Valerio-Santiago M, Monje-Casas F. The Journal of Cell Biology (2011). 192 (4): 599-614.
b) Review articles:
– “Asymmetric cell division and replicative aging: a new perspective from the spindle poles”. Manzano-López J, Monje-Casas F. Current Genetics. (2020). 66(4):719-727. doi: 10.1007/s00294-020-01074-y.
– “A guiding torch at the poles: the multiple roles of spindle microtubule-organizing centers during cell division”. Rincón AM, Monje-Casas F. Cell Cycle (2020). 19(12):1405-1421. doi: 10.1080/15384101.2020.1754586.
– “Regulation of mitotic exit by cell cycle checkpoints: Lessons from Saccharomyces cerevisiae”. Matellán L, Monje-Casas F. Genes (2020). 11(2): e195. doi.org/10.3390/genes11020195.
– “The multiple roles of the Cdc14 phosphatase in cell cycle control”. Manzano-López J, Monje-Casas F. Int. J. Mol. Sci. (2020). 21(3): e709. doi:10.3390/ijms21030709.
c) Books:
– “The Mitotic Exit Network: Methods and Protocols” (2017). Queralt E and Monje-Casas F (Editors). Book series: Methods in Molecular Biology. Volume: 1505. Publisher: Springer (New York). ISBN: 978-1-4939-6500-7 (printed version); 978-1-4939-6502-1 (online version).








