Epigenetics and gene expression
Research projects
The main goal of our group is to understand how chromatin of regulatory elements and gene bodies change during transcription, how these changes are regulated and inherited through epigenetic mechanisms and what protein factors are responsible for them. We specially investigate how alteration of these chromatin mechanisms are implicated in human disease.
- Chromatin Remodeling and Gene expression
- Enhancers and 3D chromatin structure
- Metastasis epigenetics: Epigenetic changes during epithelial to mesenchymal transition and its reversion
CHD8, a chromatin remodeler involved in autism.
Chromatin is the network where the metabolism of DNA occurs, including transcription, replication, repair and recombination. All the enzymatic machineries that have to reach the DNA for these processes require the previous concourse of chromatin remodelling complexes that open the chromatin. Some of these remodelers alter histone-DNA interactions using the energy of ATP hydrolysis. Alterations of these machineries often result in congenital malformations and cancer. Currently we are studying the role in transcriptional regulation of one of these proteins called CHD8. CHD8 binds the promoter of transcriptionally active genes and controls its expression (Rodríguez-Paredes et al., 2009; Subtil-Rodríguez et al., 2014). We have n that CHD8 is also required to activate progesterone dependent enhancers (Ceballos-Chávez et al., 2015). Recently it was discovered that heterocygotic mutations in CHD8 gene are one of the causes of autism. Currently we are investigating the molecular bases of the Autism spectrum Disorder (ASD) caused by deficiencies in this gene.
3D chromatin structure and co-expression domains (CODs)
We also study the relationship between 3D chromatin contacts and gene expression through computational methods. The study of co-transcriptional networks (Figure 1A) evidenced that coexpressed genes tend to be grouped in the genome. We call these group of collinear genes co-expression domains (CODs). Analysis of the relationship between CODs and chromatin 3D organization using Hi-C contact data, demonstrated that genes inside CODs present similar patterns of chromatin contacts (Figure 1B) (Soler-Oliva et al., 2017).
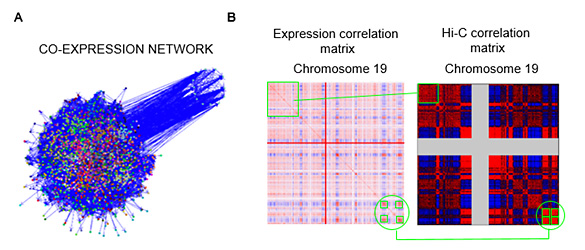
Figure 1. A. Co-expression network of breast tissue. B. Co-expression matrix of chromosome 19 (left) y correlation matrix from Hi-C chromatin contact data obtained through Hi-C (right) (PMID:19815776).
Enhancers regulation by TGFβ
Enhancers are DNA sequences that contain multiple binding sites for sequence-specific transcription factors and are classically defined as elements able of activating distant basal promoters regardless of their distance and orientation relative to them. The human genome contains more than 1.000.000 of poorly characterized enhancers which constitute an important part of the non-coding genome. Most of the diseases and predispositions caused by mutations at enhancers are uncharacterized and their study constitutes one of the major challenges of human genetics. Therefore, understanding the mechanisms of enhancer function and regulation is of paramount importance from a basic and also from a biomedical translational point of view. During the period 2018-2020 we have mapped and catalogued all the enhancers in mouse epithelial mammary cells and have characterized their regulation by the growth factor TGFβ, a trigger of EMT in epithelial cells. We have recently published that TGFβ causes a fast and widespread increase in chromatin accessibility of about 80% of all enhancers, irrespective of whether they are activated, repressed or not regulated by TGFβ (Figure 2 and 3), thus dissociating the concepts of enhancer accessibility and activity. In addition, we ed that TGFβ-regulated genes often are clustered in TGFβ regulatory domains (TRD) (Guerrero-Martínez et al., 2020).
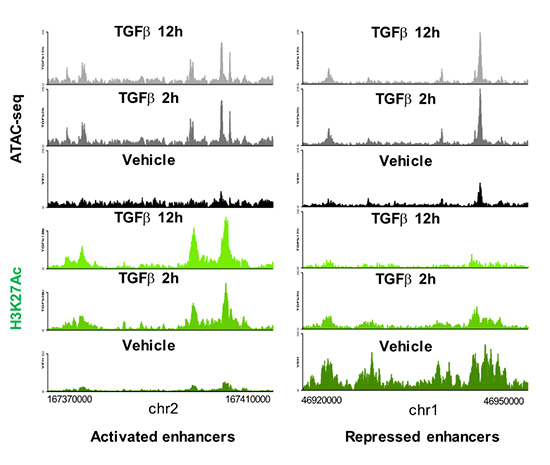
Figure 2. Enhancers landscape regulation by TGFβ.
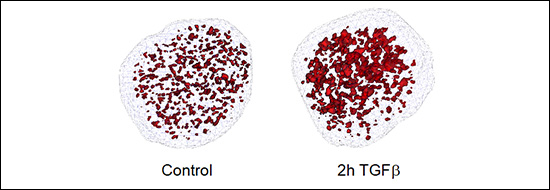
Figure 3. Open chromatin domains detected by ATAC-see after treating NMUMG cells with TGFβ.
Metastasis epigenetics: Epigenetic changes during epithelial to mesenchymal transition and its reversion.
Epithelial and mesenchymal cellular phenotypes are the edges of a spectrum of states that can be transitory (also called metastable) or stable. The epithelial to mesenchymal transition (EMT) (Figure 4) and its reversion (MET) are common processes during embryonic development and have attracted considerable interest due to the fact that they seem to be related to tumor cells dissemination and migration, generation of tumor circulating cells, cancer stem cells, chemoresistance and metastasis formation.
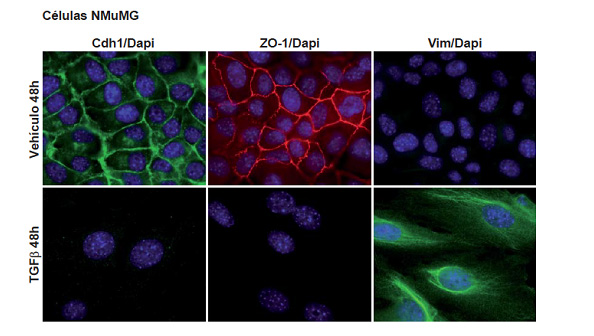
Figure 4. Epithelial to mesenchymal transition in NMuMG cell induced by TGFβ. E-Cadherin (Cdh1) (green); ZO-1 (red); Vimentin (Vim) (green) DAPI (blue).
A large reorganization of transcription patterns and epigenetic
information, that we are only starting to understand, occurs during EMT
and MET. In our group we investigate epigenetic changes that occurs
during EMT and MET and the chromatin factors implicated. We have n that
the protein HMG20A, associated to the LSD1/CoREST histone demethylase
complex, is involved in repression of epithelial genes during this
process (Rivero et al., 2015). We are studying the role of HMG20A, and
other factors identified in a genetic screen, in EMT, cancer and
metastasis. We have found that HMG20A interacts with the histone reader
PHF14 via the establishment of a two-stranded alpha-helical coiled-coil structure (Gómez-Marín et al., 2022)
Funding
- 2021-2024:
PID2020-118516GB-100 (MCIN/AEI) - 2020:2022: PY18-1962, Junta de Andalucía
- 2021:2023: PID2020-118516GB-I00. Ministerio de Ciencia, Innovación
- 2018:2020: BFU2017-85420-R, Ministerio de Ciencia, Innovación y Universidades
- 2013-present: VEC – 001/2014 FVEC-FPS. Fundación Vencer el Cáncer
Selected articles
A. Subtil-Rodríguez and J. C. Reyes. 2010. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Report. 11:751-757
Reviews








